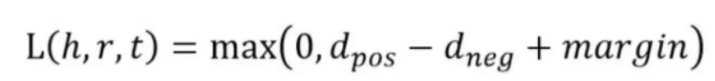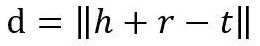模型介绍
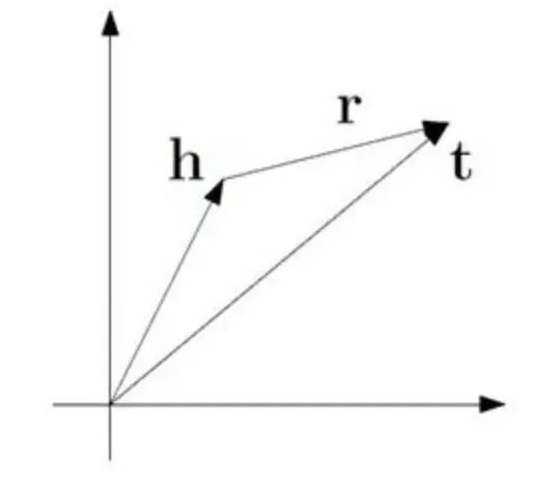
TransE模型的基本思想是使head向量和relation向量的和尽可能靠近tail向量。这里我们用L1或L2范数来衡量它们的靠近程度。
损失函数是使用了负抽样的max-margin函数。
L(y, y’) = max(0, margin – y + y’)
y是正样本的得分,y'是负样本的得分。然后使损失函数值最小化,当这两个分数之间的差距大于margin的时候就可以了(我们会设置这个值,通常是1)。
由于我们使用距离来表示得分,所以我们在公式中加上一个减号,知识表示的损失函数为:
其中,d是:
这是L1或L2范数。至于如何得到负样本,则是将head实体或tail实体替换为三元组中的随机实体。
代码实现:
具体的代码和数据集(YAGO、umls、FB15K、WN18)请见Github:
https://github.com/Colinasda/TransE.git
import codecs
import numpy as np
import copy
import time
import random
entities2id = {
}
relations2id = {
}
def dataloader(file1, file2, file3):
print("load file...")
entity = []
relation = []
with open(file2, 'r') as f1, open(file3, 'r') as f2:
lines1 = f1.readlines()
lines2 = f2.readlines()
for line in lines1:
line = line.strip().split('\t')
if len(line) != 2:
continue
entities2id[line[0]] = line[1]
entity.append(line[1])
for line in lines2:
line = line.strip().split('\t')
if len(line) != 2:
continue
relations2id[line[0]] = line[1]
relation.append(line[1])
triple_list = []
with codecs.open(file1, 'r') as f:
content = f.readlines()
for line in content:
triple = line.strip().split("\t")
if len(triple) != 3:
continue
h_ = entities2id[triple[0]]
r_ = relations2id[triple[1]]
t_ = entities2id[triple[2]]
triple_list.append([h_, r_, t_])
print("Complete load. entity : %d , relation : %d , triple : %d" % (
len(entity), len(relation), len(triple_list)))
return entity, relation, triple_list
def norm_l1(h, r, t):
return np.sum(np.fabs(h + r - t))
def norm_l2(h, r, t):
return np.sum(np.square(h + r - t))
class TransE:
def __init__(self, entity, relation, triple_list, embedding_dim=50, lr=0.01, margin=1.0, norm=1):
self.entities = entity
self.relations = relation
self.triples = triple_list
self.dimension = embedding_dim
self.learning_rate = lr
self.margin = margin
self.norm = norm
self.loss = 0.0
def data_initialise(self):
entityVectorList = {
}
relationVectorList = {
}
for entity in self.entities:
entity_vector = np.random.uniform(-6.0 / np.sqrt(self.dimension), 6.0 / np.sqrt(self.dimension),
self.dimension)
entityVectorList[entity] = entity_vector
for relation in self.relations:
relation_vector = np.random.uniform(-6.0 / np.sqrt(self.dimension), 6.0 / np.sqrt(self.dimension),
self.dimension)
relation_vector = self.normalization(relation_vector)
relationVectorList[relation] = relation_vector
self.entities = entityVectorList
self.relations = relationVectorList
def normalization(self, vector):
return vector / np.linalg.norm(vector)
def training_run(self, epochs=1, nbatches=100, out_file_title = ''):
batch_size = int(len(self.triples) / nbatches)
print("batch size: ", batch_size)
for epoch in range(epochs):
start = time.time()
self.loss = 0.0
# Normalise the embedding of the entities to 1
for entity in self.entities.keys():
self.entities[entity] = self.normalization(self.entities[entity]);
for batch in range(nbatches):
batch_samples = random.sample(self.triples, batch_size)
Tbatch = []
for sample in batch_samples:
corrupted_sample = copy.deepcopy(sample)
pr = np.random.random(1)[0]
if pr > 0.5:
# change the head entity
corrupted_sample[0] = random.sample(self.entities.keys(), 1)[0]
while corrupted_sample[0] == sample[0]:
corrupted_sample[0] = random.sample(self.entities.keys(), 1)[0]
else:
# change the tail entity
corrupted_sample[2] = random.sample(self.entities.keys(), 1)[0]
while corrupted_sample[2] == sample[2]:
corrupted_sample[2] = random.sample(self.entities.keys(), 1)[0]
if (sample, corrupted_sample) not in Tbatch:
Tbatch.append((sample, corrupted_sample))
self.update_triple_embedding(Tbatch)
end = time.time()
print("epoch: ", epoch, "cost time: %s" % (round((end - start), 3)))
print("running loss: ", self.loss)
with codecs.open(out_file_title +"TransE_entity_" + str(self.dimension) + "dim_batch" + str(batch_size), "w") as f1:
for e in self.entities.keys():
# f1.write("\t")
# f1.write(e + "\t")
f1.write(str(list(self.entities[e])))
f1.write("\n")
with codecs.open(out_file_title +"TransE_relation_" + str(self.dimension) + "dim_batch" + str(batch_size), "w") as f2:
for r in self.relations.keys():
# f2.write("\t")
# f2.write(r + "\t")
f2.write(str(list(self.relations[r])))
f2.write("\n")
def update_triple_embedding(self, Tbatch):
# deepcopy 可以保证,即使list嵌套list也能让各层的地址不同, 即这里copy_entity 和
# entitles中所有的elements都不同
copy_entity = copy.deepcopy(self.entities)
copy_relation = copy.deepcopy(self.relations)
for correct_sample, corrupted_sample in Tbatch:
correct_copy_head = copy_entity[correct_sample[0]]
correct_copy_tail = copy_entity[correct_sample[2]]
relation_copy = copy_relation[correct_sample[1]]
corrupted_copy_head = copy_entity[corrupted_sample[0]]
corrupted_copy_tail = copy_entity[corrupted_sample[2]]
correct_head = self.entities[correct_sample[0]]
correct_tail = self.entities[correct_sample[2]]
relation = self.relations[correct_sample[1]]
corrupted_head = self.entities[corrupted_sample[0]]
corrupted_tail = self.entities[corrupted_sample[2]]
# calculate the distance of the triples
if self.norm == 1:
correct_distance = norm_l1(correct_head, relation, correct_tail)
corrupted_distance = norm_l1(corrupted_head, relation, corrupted_tail)
else:
correct_distance = norm_l2(correct_head, relation, correct_tail)
corrupted_distance = norm_l2(corrupted_head, relation, corrupted_tail)
loss = self.margin + correct_distance - corrupted_distance
if loss > 0:
self.loss += loss
print(loss)
correct_gradient = 2 * (correct_head + relation - correct_tail)
corrupted_gradient = 2 * (corrupted_head + relation - corrupted_tail)
if self.norm == 1:
for i in range(len(correct_gradient)):
if correct_gradient[i] > 0:
correct_gradient[i] = 1
else:
correct_gradient[i] = -1
if corrupted_gradient[i] > 0:
corrupted_gradient[i] = 1
else:
corrupted_gradient[i] = -1
correct_copy_head -= self.learning_rate * correct_gradient
relation_copy -= self.learning_rate * correct_gradient
correct_copy_tail -= -1 * self.learning_rate * correct_gradient
relation_copy -= -1 * self.learning_rate * corrupted_gradient
if correct_sample[0] == corrupted_sample[0]:
# if corrupted_triples replaces the tail entity, the head entity's embedding need to be updated twice
correct_copy_head -= -1 * self.learning_rate * corrupted_gradient
corrupted_copy_tail -= self.learning_rate * corrupted_gradient
elif correct_sample[2] == corrupted_sample[2]:
# if corrupted_triples replaces the head entity, the tail entity's embedding need to be updated twice
corrupted_copy_head -= -1 * self.learning_rate * corrupted_gradient
correct_copy_tail -= self.learning_rate * corrupted_gradient
# normalising these new embedding vector, instead of normalising all the embedding together
copy_entity[correct_sample[0]] = self.normalization(correct_copy_head)
copy_entity[correct_sample[2]] = self.normalization(correct_copy_tail)
if correct_sample[0] == corrupted_sample[0]:
# if corrupted_triples replace the tail entity, update the tail entity's embedding
copy_entity[corrupted_sample[2]] = self.normalization(corrupted_copy_tail)
elif correct_sample[2] == corrupted_sample[2]:
# if corrupted_triples replace the head entity, update the head entity's embedding
copy_entity[corrupted_sample[0]] = self.normalization(corrupted_copy_head)
# the paper mention that the relation's embedding don't need to be normalised
copy_relation[correct_sample[1]] = relation_copy
# copy_relation[correct_sample[1]] = self.normalization(relation_copy)
self.entities = copy_entity
self.relations = copy_relation
if __name__ == '__main__':
file1 = "/umls/train.txt"
file2 = "/umls/entity2id.txt"
file3 = "/umls/relation2id.txt"
entity_set, relation_set, triple_list = dataloader(file1, file2, file3)
# modify by yourself
transE = TransE(entity_set, relation_set, triple_list, embedding_dim=30, lr=0.01, margin=1.0, norm=2)
transE.data_initialise()
transE.training_run(out_file_title="umls_")
今天的文章TransE模型的简单介绍&TransE模型的python代码实现分享到此就结束了,感谢您的阅读,如果确实帮到您,您可以动动手指转发给其他人。
版权声明:本文内容由互联网用户自发贡献,该文观点仅代表作者本人。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。如发现本站有涉嫌侵权/违法违规的内容, 请发送邮件至 举报,一经查实,本站将立刻删除。
如需转载请保留出处:https://bianchenghao.cn/25246.html