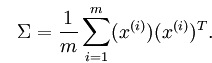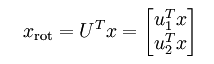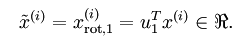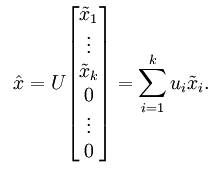一、PCA
PCA即主成分分析(Principle Components Analysis),是统计机器学习、数据挖掘中对数据进行预处理的常用的一种方法。PCA的作用有2个,一个是数据降维,一个是数据的可视化。在实际应用数据中,样本的维数可能很大,远远大于样本数目,这样模型的复杂度会很大,学习到的模型会过拟合,而且训练速度也会比较慢,内存消耗比较大,但实际数据可能有些维度是线性相关的,可能也含有噪声,这样降维处理就很有必要了,不过PCA对防止过拟合的效果也不明显,一般是根据正则化来防止过拟合。PCA是一种线性降维方法,它通过找出样本空间变化较大的一些正交的坐标方向,可认为就是样本的主成分,然后将样本投影到这些坐标从而降维到一个线性子空间。PCA找出了数据的主成分,丢掉了数据次要的成分,这些次要的成分可能是冗余或者噪声信息。PCA是一种特征选择的方法,假设数据维度为n,我们可以选取前k个方差最大的投影方向,然后把这些数据投影到这个子空间得到k维的数据,得到原数据的“压缩”表示,如果还是全部n个投影方向,我们实际只是选择了与原数据空间坐标轴方向不同的另一组坐标轴方向来表示数据而已。
PCA的两种解释分别是最大方差和最小平方误差。最大方差就是找出数据方差最大的方向,最小平方误差就是找出投影方向,使得数据投影后还是尽量地逼近原数据,跟线性回归有点类似,但实质是不一样的,线性回归是有监督的,用x值来预测y值,目的是投影后与y值误差最小,误差是样本点到直线的坐标轴距离,而PCA是无监督的,目的是要使样本与降维后的样本误差最小,误差是样本点与降维后样本点的直线距离。
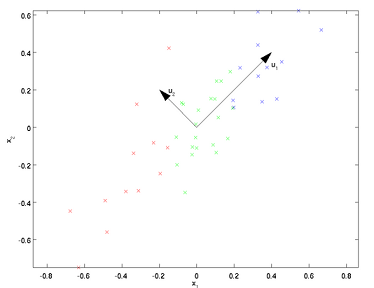
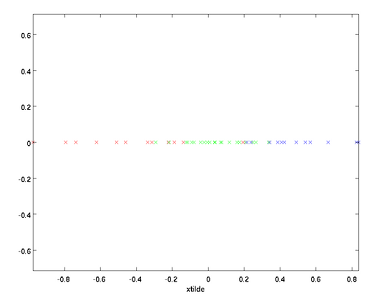
例如上图所示,u1为数据中方差最大的方向,u2为次大的方向。哪该如何找出这些投影方向呢?
首先要对数据进行零均值处理,然后求出协方差矩阵及其特征向量,特征向量就是这些投影方向。零均值处理即消减归一化、移除直流分量,要求出样本的均值,然后对每个样本点减去均值,这样,样本的每个维度的均值都为零了。协方差矩阵是一个正方矩阵,(i,j)元素表示维度i与维度j的协方差。我们的目的是要使协方差矩阵非对角上元素值为0,这样任两个不同的维度就没有相关关系了,但通过计算出来的协方差矩阵不是这样的,所以我们要找出投影方向,原数据投影后再计算的协方差矩阵的非对角元素值都为0。
协方差矩阵的计算公式为:
对协方差矩阵求特征分解求出特征向量也可以用奇异值分解(SVD)来求解,SVD求解出的U即为协方差矩阵的特征向量。U是变换矩阵或投影矩阵,即为上面说的坐标方向,U的每一列就是一个投影方向,则U’x即为x在U下的投影,如果U’取前k行(k<n),x就压缩到k维了。
特别地,对于2维空间,投影后的坐标计算如下:
投影后的数据变为:
我们可以只选择第一个投影方向(最大的特征值对应的特征向量),对数据降维:
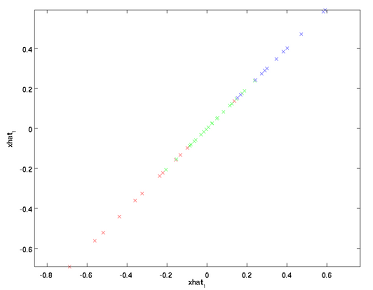
降维后的数据也可以近似还原出原来的数据,用投影矩阵U乘于降维数据尾接了n-k个0后的数据,其实就是降维时把那些方差变化小的方向的坐标值变为了0.
那如何选择k的大小,即主成分的个数呢? 一般是根据要保留多少百分比的主成分来选择k,比如可以保留99%的主成分,这样就capture了数据中的99%的成分。
二、白化
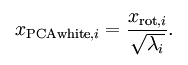
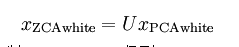
三、PCA与白化处理自然图像
http://ufldl.stanford.edu/wiki/index.php/Exercise:PCA_and_Whitening,实验数据是从自然图像中采集10000个12×12的patch。
%%================================================================
%% Step 0a: Load data
% Here we provide the code to load natural image data into x.
% x will be a 144 * 10000 matrix, where the kth column x(:, k) corresponds to
% the raw image data from the kth 12x12 image patch sampled.
% You do not need to change the code below.
x = sampleIMAGESRAW();
figure('name','Raw images');
randsel = randi(size(x,2),200,1); % A random selection of samples for visualization
display_network(x(:,randsel));
%%================================================================
%% Step 0b: Zero-mean the data (by row)
% You can make use of the mean and repmat/bsxfun functions.
% -------------------- YOUR CODE HERE --------------------
[n,m] = size(x); %m为样本数,n为样本维度
avg = mean(x, 2);
x = x - repmat(avg, 1, size(x,2));
display_network(x(:,randsel));
%%================================================================
%% Step 1a: Implement PCA to obtain xRot
% Implement PCA to obtain xRot, the matrix in which the data is expressed
% with respect to the eigenbasis of sigma, which is the matrix U.
% -------------------- YOUR CODE HERE --------------------
xRot = zeros(size(x)); % You need to compute this
sigma = x * x' / size(x, 2);
[U,S,V] = svd(sigma);
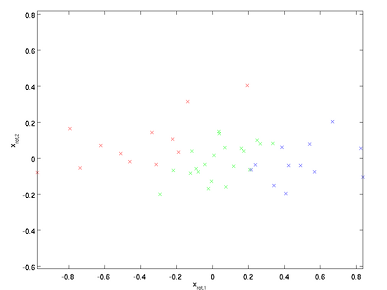
xRot = U'*x;
%%================================================================
%% Step 1b: Check your implementation of PCA
% The covariance matrix for the data expressed with respect to the basis U
% should be a diagonal matrix with non-zero entries only along the main
% diagonal. We will verify this here.
% Write code to compute the covariance matrix, covar.
% When visualised as an image, you should see a straight line across the
% diagonal (non-zero entries) against a blue background (zero entries).
% -------------------- YOUR CODE HERE --------------------
covar = zeros(size(x, 1)); % You need to compute this
covar = xRot*xRot';
% Visualise the covariance matrix. You should see a line across the
% diagonal against a blue background.
figure('name','Visualisation of covariance matrix');
imagesc(covar);
%%================================================================
%% Step 2: Find k, the number of components to retain
% Write code to determine k, the number of components to retain in order
% to retain at least 99% of the variance.
% -------------------- YOUR CODE HERE --------------------
k = 0; % Set k accordingly
var_sum = sum(diag(covar));
curr_var_sum = 0;
for i=1:length(covar)
curr_var_sum = curr_var_sum + covar(i,i);
if curr_var_sum / var_sum >= 0.99
k = i;
break
end
end
%%================================================================
%% Step 3: Implement PCA with dimension reduction
% Now that you have found k, you can reduce the dimension of the data by
% discarding the remaining dimensions. In this way, you can represent the
% data in k dimensions instead of the original 144, which will save you
% computational time when running learning algorithms on the reduced
% representation.
%
% Following the dimension reduction, invert the PCA transformation to produce
% the matrix xHat, the dimension-reduced data with respect to the original basis.
% Visualise the data and compare it to the raw data. You will observe that
% there is little loss due to throwing away the principal components that
% correspond to dimensions with low variation.
% -------------------- YOUR CODE HERE --------------------
xHat = zeros(size(x)); % You need to compute this
xTilde = U(:, 1:k)'*x;
xHat = U*[xTilde; zeros(n-k,m)];
% Visualise the data, and compare it to the raw data
% You should observe that the raw and processed data are of comparable quality.
% For comparison, you may wish to generate a PCA reduced image which
% retains only 90% of the variance.
figure('name',['PCA processed images ',sprintf('(%d / %d dimensions)', k, size(x, 1)),'']);
display_network(xHat(:,randsel));
figure('name','Raw images');
display_network(x(:,randsel));
%%================================================================
%% Step 4a: Implement PCA with whitening and regularisation
% Implement PCA with whitening and regularisation to produce the matrix
% xPCAWhite.
epsilon = 1;
xPCAWhite = zeros(size(x));
xPCAWhite = diag(1./sqrt(diag(S)+epsilon)) * xRot;
% -------------------- YOUR CODE HERE --------------------
%%================================================================
%% Step 4b: Check your implementation of PCA whitening
% Check your implementation of PCA whitening with and without regularisation.
% PCA whitening without regularisation results a covariance matrix
% that is equal to the identity matrix. PCA whitening with regularisation
% results in a covariance matrix with diagonal entries starting close to
% 1 and gradually becoming smaller. We will verify these properties here.
% Write code to compute the covariance matrix, covar.
%
% Without regularisation (set epsilon to 0 or close to 0),
% when visualised as an image, you should see a red line across the
% diagonal (one entries) against a blue background (zero entries).
% With regularisation, you should see a red line that slowly turns
% blue across the diagonal, corresponding to the one entries slowly
% becoming smaller.
% -------------------- YOUR CODE HERE --------------------
covar = xPCAWhite*xPCAWhite';
% Visualise the covariance matrix. You should see a red line across the
% diagonal against a blue background.
figure('name','Visualisation of covariance matrix');
imagesc(covar);
%%================================================================
%% Step 5: Implement ZCA whitening
% Now implement ZCA whitening to produce the matrix xZCAWhite.
% Visualise the data and compare it to the raw data. You should observe
% that whitening results in, among other things, enhanced edges.
xZCAWhite = zeros(size(x));
xZCAWhite = U'*xPCAWhite;
% -------------------- YOUR CODE HERE --------------------
% Visualise the data, and compare it to the raw data.
% You should observe that the whitened images have enhanced edges.
figure('name','ZCA whitened images');
display_network(xZCAWhite(:,randsel));
figure('name','Raw images');
display_network(x(:,randsel));
版权声明:本文内容由互联网用户自发贡献,该文观点仅代表作者本人。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。如发现本站有涉嫌侵权/违法违规的内容, 请发送邮件至 举报,一经查实,本站将立刻删除。
如需转载请保留出处:https://bianchenghao.cn/10797.html