libsvm是基于支持向量机(support vector machine, SVM)实现的开源库,由台湾大学林智仁(Chih-Jen Lin)教授等开发,它主要用于分类(支持二分类和多分类)和回归。它的License是BSD-3-Clause,最新发布版本是v322。libsvm具有操作简单、易于使用、快速有效、且对SVM所涉及的参数调节相对较少的特点。Code地址: https://github.com/cjlin1/libsvm
libsvm直接支持的开发语言包括:C++、java、matlab、python。C++主要包括两个文件,一个是头文件svm.h,一个是实现文件svm.cpp,将这两个文件加入到工程中就可以使用libsvm了。
在windows平台下,可以直接使用libsvm/windows目录下已经编译好的执行文件。
关于支持向量机的基础介绍可以参考: libsvm库简介及使用
libsvm中支持的svm类型包括:C-SVC(multi-class classification)、nu-SVC(multi-class classification)、one-class SVM、epsilon-SVR(regression)、nu-SVR(regression);支持的kernel类型包括:linear(u’*v)、polynomial((gamma*u’*v+coef0)^degree)、radial basis function(径向基, exp(-gamma*|u-v|^2))、sigmoid(tanh(gamma*u’*v+coef0))、precomputed kernel(kernel values in training_set_file)。libsvm中需要的参数虽然较多,但是绝大多数都有其默认值。
libsvm中对数据集进行缩放的目的在于:避免一些特征值范围过大而另一些特征值范围过小;避免在训练时为了计算核函数而计算内积的时候引起数值计算的困难。通常将数据缩放到[-1, 1]或[0, 1]之间。
libsvm使用一般步骤:
(1)、按照libsvm所要求的格式准备数据集;
(2)、对数据进行简单的缩放操作;
(3)、考虑选用RBF(radial basis function)核参数;
(4)、如果选用RBF,通过libsvm/tools/grid.py采用交叉验证获取最佳参数C与gamma;
(5)、采用最佳参数C与g对整个训练集进行训练获取支持向量机模型;
(6)、利用获取的模型进行测试与预测。
测试代码执行步骤:
(1)、训练和测试数据采用UCI/Liver-disorders( https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html#liver-disorders ):二分类、训练样本为145、测试样本为200、特征长度为5,分别保存为liver-disorders_train.txt、liver-disorders_predict.txt;
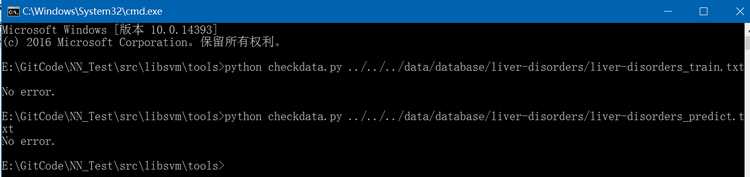
(2)、通过libsvm/tools/checkdata.py来验证liver-disorders数据集是否满足libsvm所要求的数据格式(注:在windows下,此两个.txt文件编码格式要由utf-8改为ansi,否则执行checkdata.py会有errror),执行结果如下:
(3)、调用test_libsvm_scale函数对liver-disorders_train.txt进行缩放操作,并保存scale后的数据liver-disorders_train_scale.txt,并通过checkdata.py验证数据格式;
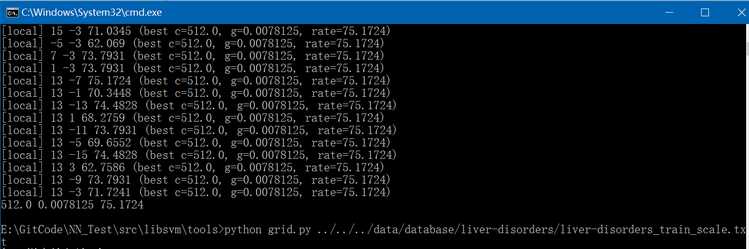
(4)、通过liver-disorders_train_scale.txt文件调用libsvm/tools/grid.py生成最优参数C和gamma,执行结果如下:
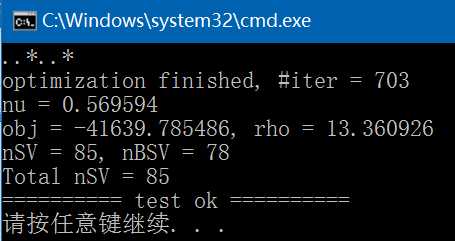
(5)、train,生成svm.model文件,执行结果如下,与libsvm/windows/svm-train.exe执行结果完全一致:
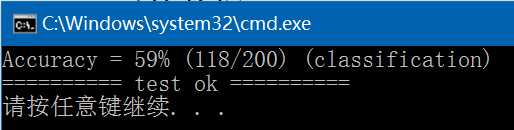
(6)、predict,执行结果如下,与libsvm/windows/svm-predict.exe执行结果完全一致:
以下二分类测试代码是参考libsvm中的svm-scale.c、svm-train.c、svm-predict.c:
#include "libsvm.hpp"
#include <iostream>
#include <limits>
#include <algorithm>
#include <fstream>
#include "svm.h"
#include "common.hpp"
#define Malloc(type,n) (type *)malloc((n)*sizeof(type))
static char* readline(FILE* input, int& max_line_len, char** line)
{
if (fgets(*line, max_line_len, input) == nullptr) return nullptr;
int len{ 0 };
while (strrchr(*line, '\n') == nullptr) {
max_line_len *= 2;
*line = (char *)realloc(*line, max_line_len);
len = (int)strlen(*line);
if (fgets(*line + len, max_line_len - len, input) == nullptr) break;
}
return *line;
}
static void output(const double* feature_max, const double* feature_min, double lower, double upper, long int& new_num_nonzeros, int index, double value, std::ofstream& out_file)
{
/* skip single-valued attribute */
if (feature_max[index] == feature_min[index]) return;
if (value == feature_min[index]) value = lower;
else if (value == feature_max[index]) value = upper;
else value = lower + (upper - lower) * (value - feature_min[index]) / (feature_max[index] - feature_min[index]);
if (value != 0) {
//fprintf(stdout, "%d:%g ", index, value);
out_file << index << ":" << value<<" ";
new_num_nonzeros++;
}
}
int test_libsvm_scale(const char* input_file_name, const char* output_file_name)
{
// reference: libsvm/svm-scale.c
const double lower{ -1. }, upper{ 1. }; // lower: x scaling lower limit(default -1); upper: x scaling upper limit(default +1)
const double y_lower{ 0. }, y_upper{ 1. }, y_scaling{ 0. }; // y scaling limits (default: no y scaling)
double y_max{ std::numeric_limits<double>::max() };
double y_min{ std::numeric_limits<double>::lowest() };
char* line{ nullptr };
int max_line_len{ 1024 };
double* feature_max{ nullptr };
double* feature_min{ nullptr };
int max_index{ 0 }, min_index{ 0 }, index{ 0 };
long int num_nonzeros{ 0 }, new_num_nonzeros{ 0 };
FILE* fp = fopen(input_file_name, "r");
CHECK(nullptr != fp);
line = (char *)malloc(max_line_len * sizeof(char));
#define SKIP_TARGET \
while (isspace(*p)) ++p; \
while (!isspace(*p)) ++p;
#define SKIP_ELEMENT \
while (*p != ':') ++p; \
++p; \
while (isspace(*p)) ++p; \
while (*p && !isspace(*p)) ++p;
// pass 1: find out max index of attributes
max_index = 0;
min_index = 1;
while (readline(fp, max_line_len, &line) != nullptr) {
char* p = line;
SKIP_TARGET
while (sscanf(p, "%d:%*f", &index) == 1) {
max_index = std::max(max_index, index);
min_index = std::min(min_index, index);
SKIP_ELEMENT
num_nonzeros++;
}
}
CHECK(min_index >= 1);
rewind(fp);
feature_max = (double *)malloc((max_index + 1)* sizeof(double));
feature_min = (double *)malloc((max_index + 1)* sizeof(double));
CHECK(feature_max != nullptr && feature_min != nullptr);
for (int i = 0; i <= max_index; ++i) {
feature_max[i] = std::numeric_limits<double>::lowest();
feature_min[i] = std::numeric_limits<double>::max();
}
// pass 2: find out min/max value
while (readline(fp, max_line_len, &line) != nullptr) {
char *p = line;
int next_index = 1;
double target;
double value;
CHECK(sscanf(p, "%lf", &target) == 1);
y_max = std::max(y_max, target);
y_min = std::min(y_min, target);
SKIP_TARGET
while (sscanf(p, "%d:%lf", &index, &value) == 2) {
for (int i = next_index; i<index; ++i) {
feature_max[i] = std::max(feature_max[i], 0.);
feature_min[i] = std::min(feature_min[i], 0.);
}
feature_max[index] = std::max(feature_max[index], value);
feature_min[index] = std::min(feature_min[index], value);
SKIP_ELEMENT
next_index = index + 1;
}
for (int i = next_index; i <= max_index; ++i) {
feature_max[i] = std::max(feature_max[i], 0.);
feature_min[i] = std::min(feature_min[i], 0.);
}
}
rewind(fp);
std::ofstream out_file(output_file_name);
CHECK(out_file);
// pass 3: scale
while (readline(fp, max_line_len, &line) != nullptr) {
char *p = line;
int next_index = 1;
double target;
double value;
CHECK(sscanf(p, "%lf", &target) == 1);
//fprintf(stdout, "%g ", target);
out_file << target << " ";
SKIP_TARGET
while (sscanf(p, "%d:%lf", &index, &value) == 2) {
for (int i = next_index; i<index; ++i)
output(feature_max, feature_min, lower, upper, new_num_nonzeros, i, 0, out_file);
output(feature_max, feature_min, lower, upper, new_num_nonzeros, index, value, out_file);
SKIP_ELEMENT
next_index = index + 1;
}
for (int i = next_index; i <= max_index; ++i)
output(feature_max, feature_min, lower, upper, new_num_nonzeros, i, 0, out_file);
//fprintf(stdout, "\n");
out_file << std::endl;
}
CHECK(new_num_nonzeros <= num_nonzeros);
free(line);
free(feature_max);
free(feature_min);
fclose(fp);
out_file.close();
return 0;
}
static int read_problem(const char* input_file_name, svm_problem& prob, int& max_line_len, svm_parameter& param, char** line, svm_node** x_space)
{
int max_index, inst_max_index;
size_t elements, j;
char* endptr;
char *idx, *val, *label;
FILE* fp = fopen(input_file_name, "r");
CHECK(fp != nullptr);
prob.l = 0;
elements = 0;
max_line_len = 1024;
*line = Malloc(char, max_line_len);
while (readline(fp, max_line_len, line) != nullptr) {
char *p = strtok(*line, " \t"); // label
// features
while (1) {
p = strtok(nullptr, " \t");
if (p == nullptr || *p == '\n') // check '\n' as ' ' may be after the last feature
break;
++elements;
}
++elements;
++prob.l;
}
rewind(fp);
prob.y = Malloc(double, prob.l);
prob.x = Malloc(struct svm_node *, prob.l);
*x_space = Malloc(struct svm_node, elements);
max_index = 0;
j = 0;
for (int i = 0; i<prob.l; i++) {
inst_max_index = -1; // strtol gives 0 if wrong format, and precomputed kernel has <index> start from 0
readline(fp, max_line_len, line);
prob.x[i] = &(*x_space)[j];
label = strtok(*line, " \t\n");
CHECK(label != nullptr); // empty line
prob.y[i] = strtod(label, &endptr);
CHECK(!(endptr == label || *endptr != '\0'));
while (1) {
idx = strtok(nullptr, ":");
val = strtok(nullptr, " \t");
if (val == nullptr) break;
errno = 0;
(*x_space)[j].index = (int)strtol(idx, &endptr, 10);
if (endptr == idx || errno != 0 || *endptr != '\0' || (*x_space)[j].index <= inst_max_index) {
CHECK(0);
} else {
inst_max_index = (*x_space)[j].index;
}
errno = 0;
(*x_space)[j].value = strtod(val, &endptr);
if (endptr == val || errno != 0 || (*endptr != '\0' && !isspace(*endptr))) CHECK(0);
++j;
}
if (inst_max_index > max_index) max_index = inst_max_index;
(*x_space)[j++].index = -1;
}
if (param.gamma == 0 && max_index > 0) param.gamma = 1.0 / max_index;
if (param.kernel_type == PRECOMPUTED) {
for (int i = 0; i<prob.l; i++) {
CHECK(prob.x[i][0].index == 0);
if ((int)prob.x[i][0].value <= 0 || (int)prob.x[i][0].value > max_index) {
CHECK(0);
}
}
}
fclose(fp);
return 0;
}
int test_libsvm_two_classification_train()
{
// reference: libsvm/svm-train.c
const std::string input_file_name{ "E:/GitCode/NN_Test/data/database/liver-disorders/liver-disorders_train.txt" },
output_file_name{ "E:/GitCode/NN_Test/data/database/liver-disorders/liver-disorders_train_scale.txt" },
svm_model{"E:/GitCode/NN_Test/data/svm.model"};
CHECK(0 == test_libsvm_scale(input_file_name.c_str(), output_file_name.c_str())); // data scale
struct svm_parameter param = {};
struct svm_problem prob = {};
struct svm_model* model = nullptr;
struct svm_node* x_space = nullptr;
int cross_validation{ 0 };
int nr_fold{0};
char* line = nullptr;
int max_line_len{0};
param = { C_SVC, RBF, 3, 0.0078125, 0., 100, 1e-3, 512., 0, nullptr, nullptr, 0.5, 0.1, 1, 0 };
CHECK(read_problem(output_file_name.c_str(), prob, max_line_len, param, &line, &x_space) == 0);
CHECK(svm_check_parameter(&prob, ¶m) == nullptr);
model = svm_train(&prob, ¶m);
CHECK(svm_save_model(svm_model.c_str(), model) == 0);
svm_free_and_destroy_model(&model);
svm_destroy_param(¶m);
free(prob.y);
free(prob.x);
free(x_space);
free(line);
return 0;
}
static int predict(FILE* input, FILE* output, const svm_model* model, int& max_line_len, char** line, int& max_nr_attr, svm_node** x)
{
int correct = 0;
int total = 0;
double error = 0;
double sump = 0, sumt = 0, sumpp = 0, sumtt = 0, sumpt = 0;
int svm_type = svm_get_svm_type(model);
int nr_class = svm_get_nr_class(model);
double *prob_estimates = nullptr;
int j;
max_line_len = 1024;
*line = (char *)malloc(max_line_len*sizeof(char));
while (readline(input, max_line_len, line) != nullptr) {
int i = 0;
double target_label, predict_label;
char *idx, *val, *label, *endptr;
int inst_max_index = -1; // strtol gives 0 if wrong format, and precomputed kernel has <index> start from 0
label = strtok(*line, " \t\n");
CHECK(label != nullptr); // empty line
target_label = strtod(label, &endptr);
CHECK(!(endptr == label || *endptr != '\0'));
while (1) {
if (i >= max_nr_attr - 1) { // need one more for index = -1
max_nr_attr *= 2;
*x = (struct svm_node *) realloc(*x, max_nr_attr*sizeof(struct svm_node));
}
idx = strtok(nullptr, ":");
val = strtok(nullptr, " \t");
if (val == nullptr) break;
errno = 0;
(*x)[i].index = (int)strtol(idx, &endptr, 10);
CHECK(!(endptr == idx || errno != 0 || *endptr != '\0' || (*x)[i].index <= inst_max_index));
inst_max_index = (*x)[i].index;
errno = 0;
(*x)[i].value = strtod(val, &endptr);
CHECK(!(endptr == val || errno != 0 || (*endptr != '\0' && !isspace(*endptr))));
++i;
}
(*x)[i].index = -1;
predict_label = svm_predict(model, *x);
fprintf(output, "%g\n", predict_label);
if (predict_label == target_label) ++correct;
error += (predict_label - target_label)*(predict_label - target_label);
sump += predict_label;
sumt += target_label;
sumpp += predict_label*predict_label;
sumtt += target_label*target_label;
sumpt += predict_label*target_label;
++total;
}
fprintf(stdout, "Accuracy = %g%% (%d/%d) (classification)\n", (double)correct / total * 100, correct, total);
return 0;
}
int test_libsvm_two_classification_predict()
{
// reference: libsvm/svm-predict.c
const std::string input_file_name{ "E:/GitCode/NN_Test/data/database/liver-disorders/liver-disorders_predict.txt" },
scale_file_name{ "E:/GitCode/NN_Test/data/database/liver-disorders/liver-disorders_predict_scale.txt" },
svm_model{ "E:/GitCode/NN_Test/data/svm.model" },
predict_result_file_name{ "E:/GitCode/NN_Test/data/svm_predict_result.txt" };
CHECK(0 == test_libsvm_scale(input_file_name.c_str(), scale_file_name.c_str())); // data scale
struct svm_node* x = nullptr;
int max_nr_attr = 64;
struct svm_model* model = nullptr;
int predict_probability = 0;
char* line = nullptr;
int max_line_len = 0;
FILE* input = fopen(scale_file_name.c_str(), "r");
CHECK(input != nullptr);
FILE* output = fopen(predict_result_file_name.c_str(), "w");
CHECK(output != nullptr);
CHECK((model = svm_load_model(svm_model.c_str())) != nullptr);
x = (struct svm_node *) malloc(max_nr_attr*sizeof(struct svm_node));
CHECK(svm_check_probability_model(model) == 0);
predict(input, output, model, max_line_len, &line, max_nr_attr, &x);
svm_free_and_destroy_model(&model);
free(x);
free(line);
fclose(input);
fclose(output);
return 0;
}
GitHub:
https://github.com/fengbingchun/NN_Test
版权声明:本文内容由互联网用户自发贡献,该文观点仅代表作者本人。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。如发现本站有涉嫌侵权/违法违规的内容, 请发送邮件至 举报,一经查实,本站将立刻删除。
如需转载请保留出处:https://bianchenghao.cn/76328.html