RagTag简介
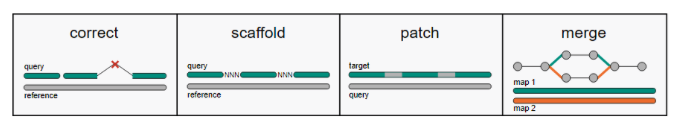
RagTag可以进行错误组装校正、scaffold组装和修补、scaffold合并等,一共分四步:correct,scaffold,patch,merge。之后,可以用Liftoff进行基因注释。
RagTag的conda安装
conda install -c bioconda ragtag1. correct
校正是使用参考基因组来鉴定和校正contigs中的组装错误,该步骤不会将序列减少或增加,仅仅是将序列在错误组装的位置进行打断。
ragtag.py correct relatives-reference.fa scaffolds_FINAL.fasta -t 80结果输出在文件夹./ragtag_output/中。
所有参数:
usage: ragtag.py correct <reference.fa> <query.fa>
Homology-based misassembly correction: Correct sequences in 'query.fa' by comparing them to sequences in 'reference.fa'>
positional arguments:
<reference.fa> reference fasta file (uncompressed or bgzipped)
<query.fa> query fasta file (uncompressed or bgzipped)
optional arguments:
-h, --help show this help message and exit
correction options:
-f INT minimum unique alignment length [1000]
--remove-small remove unique alignments shorter than -f
-q INT minimum mapq (NA for Nucmer alignments) [10]
-d INT maximum alignment merge distance [100000]
-b INT minimum break distance from contig ends [5000]
-e <exclude.txt> list of reference headers to ignore [null]
-j <skip.txt> list of query headers to leave uncorrected [null]
--inter only break misassemblies between reference sequences
--intra only break misassemblies within reference sequences
--gff <features.gff> don't break sequences within gff intervals [null]
input/output options:
-o PATH output directory [./ragtag_output]
-w overwrite intermediate files
-u add suffix to unaltered sequence headers
mapping options:
-t INT number of minimap2/unimap threads [1]
--aligner PATH whole genome aligner executable ('nucmer', 'unimap' or 'minimap2') [minimap2]
--mm2-params STR space delimited minimap2 whole genome alignment parameters (overrides '-t') ['-x asm5']
--unimap-params STR space delimited unimap parameters (overrides '-t') ['-x asm5']
--nucmer-params STR space delimted nucmer whole genome alignment parameters ['--maxmatch -l 100 -c 500']
validation options:
--read-aligner PATH read aligner executable (only 'minimap2' is allowed) [minimap2]
-R <reads.fasta> validation reads (uncompressed or gzipped) [null]
-F <reads.fofn> same as '-R', but a list of files [null]
-T STR read type. 'sr', 'ont' and 'corr' accepted for Illumina, nanopore and error corrected long-reads,
respectively [null]
-v INT coverage validation window size [10000]
--max-cov INT break sequences at regions at or above this coverage level [AUTO]
--min-cov INT break sequences at regions at or below this coverage level [AUTO]
** The reference and query FASTA files are required **2. scaffold
该步骤是将相邻的contigs序列用100个N连起来,序列的位置和方向需要根据与参考基因组的比对结果确定。
ragtag.py scaffold relatives-reference.fa ragtag_output/ragtag.correct.fasta -t 80 -C ## -C会将没地方放的contig/scaffold连在一起放到chr0中(中间用100个N连接)
最后得到的重要的结果:
ragtag.scaffold.fasta
ragtag.scaffold.agp
插播:
如果除了chr0还有一些碎的contig没放进去,可以用下面的python脚本处理:
#!/public/home/zhangchaofan/anaconda3/bin/python
# -*- coding: utf-8 -*-
##python 02.add_seq.py -s mapped.filter.list.fasta -g test.apg -o test.fa
"""
@File : add_seq.py
@Time : 2021-12-14 22:07:37
@Version : 1.0
@License : (C)Copyright 2020-2021
@Desc : None
@Usage : None
"""
import argparse
from multiprocessing import Pool
import os
import subprocess
import sys
def err_exit():
sys.exit('[1;31;47m!!The program exited abnormally, please check the log file !![0m')
def Argparse():
group = argparse.ArgumentParser()
group.add_argument("-i", '--inf', help="Please input total_seq_file!")
group.add_argument('-s', '--seqs', help="Please input the to_add_seq")
group.add_argument('-g', '--gff', help="please input the gff_file!")
return group.parse_args()
def main():
inf_lines = open(inf).readlines()
total_seq = inf_lines[1].strip()
# 将要添加的序列读入内存
id_seq = {}
for line in open(seqs).readlines():
if line[0] == '>':
# 不保留 >
id = line.strip()[1:]
else:
id_seq[id] = line.strip()
sort_id = sorted(id_seq.items(), key=lambda item: len(item[1]), reverse=True)
# end_pos = 134326222
# 先加N 后加序列
gff_lines = open(gff).readlines()
end_temp = gff_lines[-1].strip().split()
end_pos = int(end_temp[2])
end_order = int(end_temp[3]) + 1
for tuple_ in sort_id:
total_seq += 'N' * 100
temp_gff_line = 'Chr0_RagTag\t%d\t%d\t%d\tU\t100\tcontig\tno\tna\n' % (end_pos+1, end_pos+100, end_order)
end_order += 1
gff_lines.append(temp_gff_line)
total_seq += tuple_[1]
end_pos += 100
temp_gff_line = 'Chr0_RagTag\t%d\t%d\t%d\tW\t%s\t1\t%d\t+\n' % (end_pos+1, end_pos+len(tuple_[1]), end_order, tuple_[0], len(tuple_[1]))
gff_lines.append(temp_gff_line)
end_order += 1
end_pos += len(tuple_[1])
gff_ouf_w = open(gff+'.change', 'w')
gff_ouf_w.write(''.join(gff_lines))
gff_ouf_w.close()
seq_ouf = open(inf+'.change', 'w')
seq_ouf.write(inf_lines[0]+total_seq+'\n')
seq_ouf.close()
if __name__ == '__main__':
opts = Argparse()
inf = opts.inf
seqs = opts.seqs
gff = opts.gff
main()
chr0.list.fasta ##已合并chr0的fasta序列文件
contig.list.fasta ##剩下还有的contig的fasta序列文件
运行后contig.list.fasta中的序列会从大到小接在chr0后面,中间用100个N连接,最后生成新的fasta文件和agp文件。
所有参数:
usage: ragtag.py scaffold <reference.fa> <query.fa>
Homology-based assembly scaffolding: Order and orient sequences in 'query.fa' by comparing them to sequences in 'reference.fa'
positional arguments:
<reference.fa> reference fasta file (uncompressed or bgzipped)
<query.fa> query fasta file (uncompressed or bgzipped)
optional arguments:
-h, --help show this help message and exit
scaffolding options:
-e <exclude.txt> list of reference sequences to ignore [null]
-j <skip.txt> list of query sequences to leave unplaced [null]
-J <hard-skip.txt> list of query headers to leave unplaced and exclude from 'chr0' ('-C') [null]
-f INT minimum unique alignment length [1000]
--remove-small remove unique alignments shorter than '-f'
-q INT minimum mapq (NA for Nucmer alignments) [10]
-d INT maximum alignment merge distance [100000]
-i FLOAT minimum grouping confidence score [0.2]
-a FLOAT minimum location confidence score [0.0]
-s FLOAT minimum orientation confidence score [0.0]
-C concatenate unplaced contigs and make 'chr0'
-r infer gap sizes. if not, all gaps are 100 bp
-g INT minimum inferred gap size [100]
-m INT maximum inferred gap size [100000]
input/output options:
-o PATH output directory [./ragtag_output]
-w overwrite intermediate files
-u add suffix to unplaced sequence headers
mapping options:
-t INT number of minimap2/unimap threads [1]
--aligner PATH aligner executable ('nucmer', 'unimap' or 'minimap2') [minimap2]
--mm2-params STR space delimited minimap2 parameters (overrides '-t') ['-x asm5']
--unimap-params STR space delimited unimap parameters (overrides '-t') ['-x asm5']
--nucmer-params STR space delimted nucmer parameters ['--maxmatch -l 100 -c 500']
** The reference and query FASTA files are required **3. patch
该步骤是用contigs序列对上一步得到的scaffold序列进行gap填补。该步骤比较耗时,如果急需使用基因组进行后续分析,可以省略该步骤。
ragtag.py patch ./ragtag_output/ragtag.scaffold.fasta scaffolds_FINAL.fasta -t 80以上均已亲测。
4. merge
在scaffolding过程中,可能会根据不同参数或图谱数据产生多个版本的基因组组装结果,该步骤可以将多个结果根据权重进行最终组装结果的生成。
如果有HiC数据,还可以加入HiC数据生成比较好的组装结果。
参考来源:
基于参考基因组的基因组组装和注释 – 简书
GitHub – malonge/RagTag: Tools for fast and flexible genome assembly scaffolding and improvement
今天的文章同源基因构建基因进化树_GATA2突变是先天的吗分享到此就结束了,感谢您的阅读。
版权声明:本文内容由互联网用户自发贡献,该文观点仅代表作者本人。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。如发现本站有涉嫌侵权/违法违规的内容, 请发送邮件至 举报,一经查实,本站将立刻删除。
如需转载请保留出处:https://bianchenghao.cn/89684.html